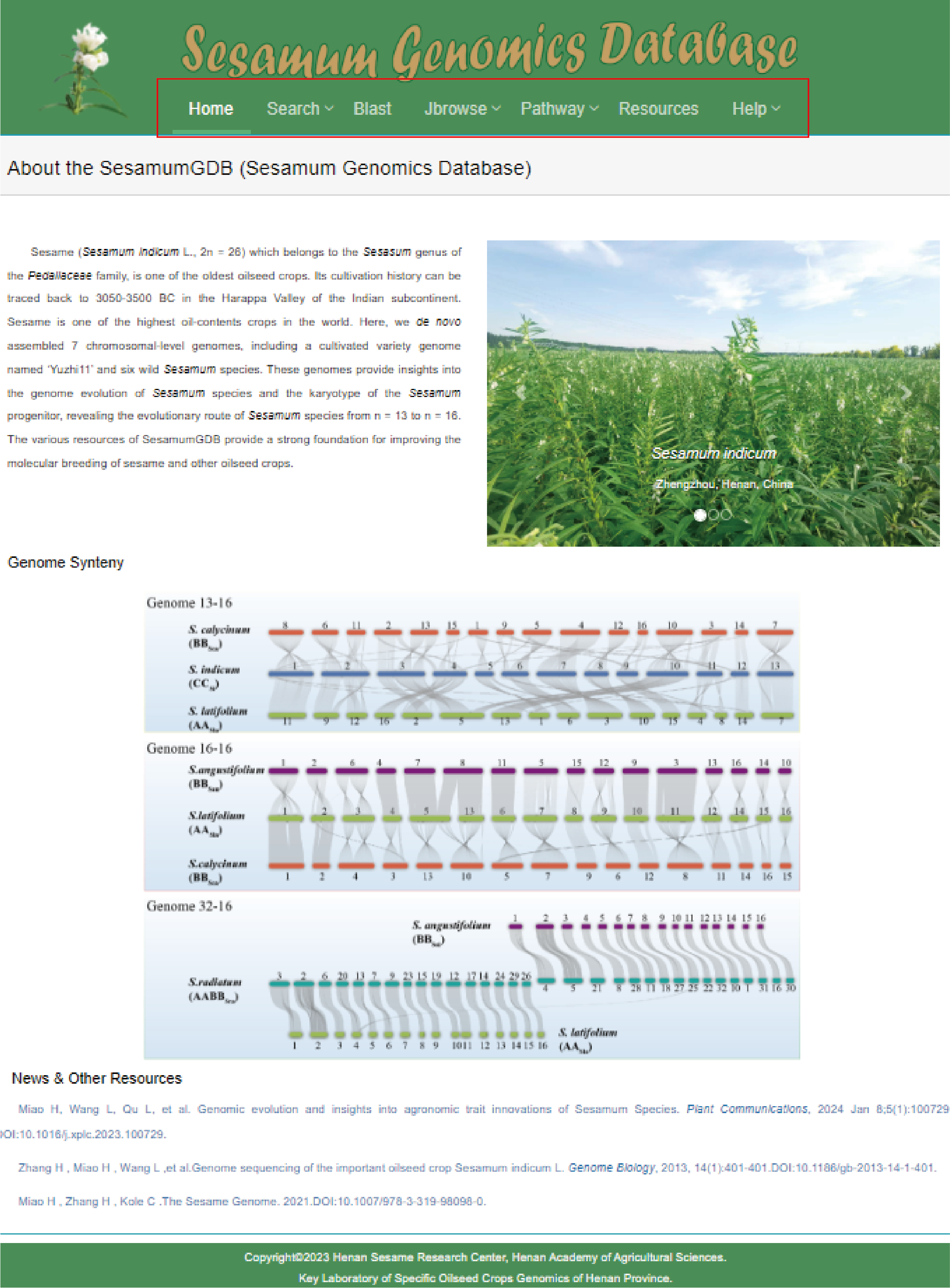
SesamumGDB offers a suite of user-friendly features, including Search, Browse, Blast, and Download. To ensure ease of use, a comprehensive help page has been developed.
The homepage is displayed in the following:
Main functions of the database are provided in menu bar form (boxed in red). Users can simply click on the menu item that piques their interest to be directed to the corresponding web page
Both gene, lipid genes, gene families, and transcription factors were suported
The Gene Retrieve page:
1. The database offers two distinct search categories for genes: 'Species' selection through a dropdown menu and direct 'Gene ID' entry.
2. Under the 'Species' category, users have access to a selection of eight species within the database.

The Lipid Genes page:
1. The database features an advanced lipid genes search functionality with two primary categories:
Species: Easily select from a dropdown menu of eight distinct species available in our database.
Sub-processes of Lipid Metabolism: Dive deeper with nine specialized search entries, covering a range of lipid metabolic activities including:
lipid-A biosynthesis;
fatty acid metabolism;
cytoplasmic lipid droplet-associated activities;
galactolipid and sulfolipid biosynthesis;
glycerolipid metabolism;
lipid trafficking;
phytosterol metabolism;
plastoglobule-associated activities;
sphingolipid metabolism.
2. Under the 'Species' category, users have access to a selection of eight species within the database.

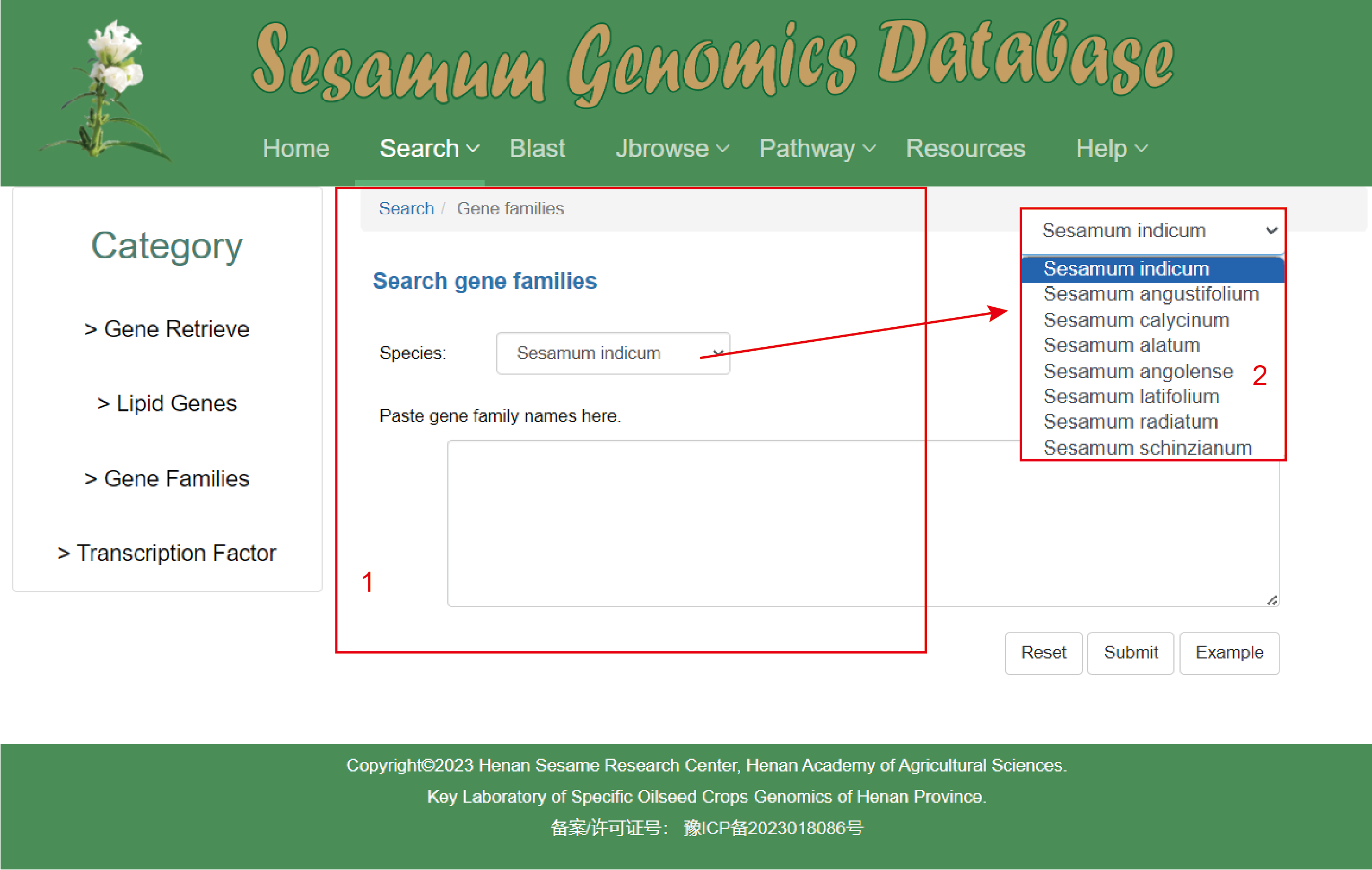
The Gene Family page:
1. The database offers two distinct search categories for genes: 'Species' selection through a dropdown menu and direct gene family names entry.
2. Under the 'Species' category, users have access to a selection of eight species within the database.
The Transcription Factor page:
The database provides a multi-search function for TFs, allowing users to directly enter gene ID, species names, variety names, and TF names.

Homologus BLAST
1. Users may paste their query sequence(s) directly into the input field or utilize the drag-and-drop feature to upload a FASTA-formatted file into the designated area.
2.Choose the database (nucleotide or protein) to be used for homologue BLAST analysis.
3. Users can opt for the default settings or customize the analysis parameters through the 'Advanced Options' interface, such as e-value, scoring matrix, and output formatting.

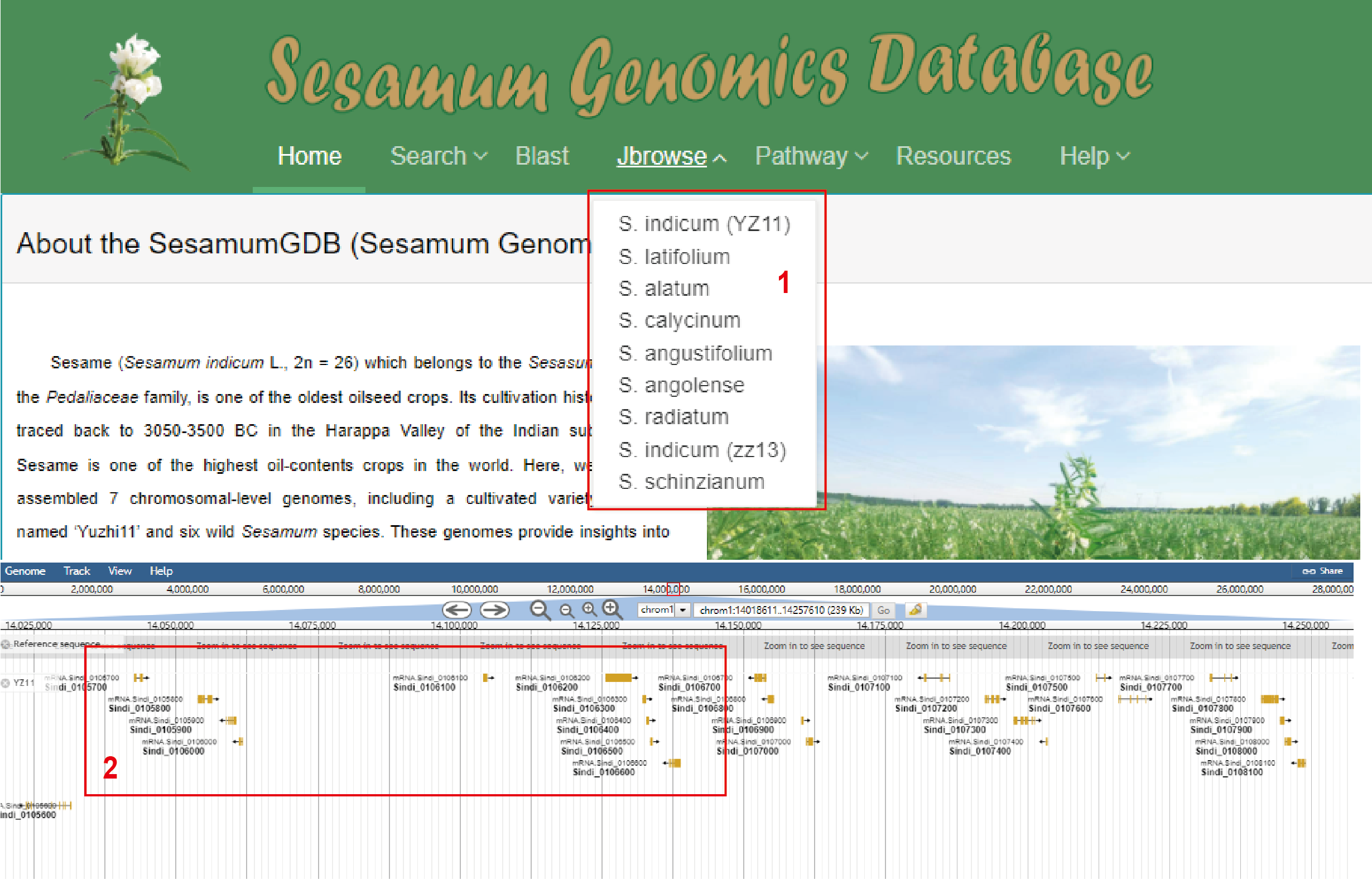
1. The genome browser of JBrowse has been constructed for the nine high-quality Sesamum genomes, enabling the viewing and searching of the genome sequence by gene IDs or genomic locations.
2. Click the icon to access detailed gene information.
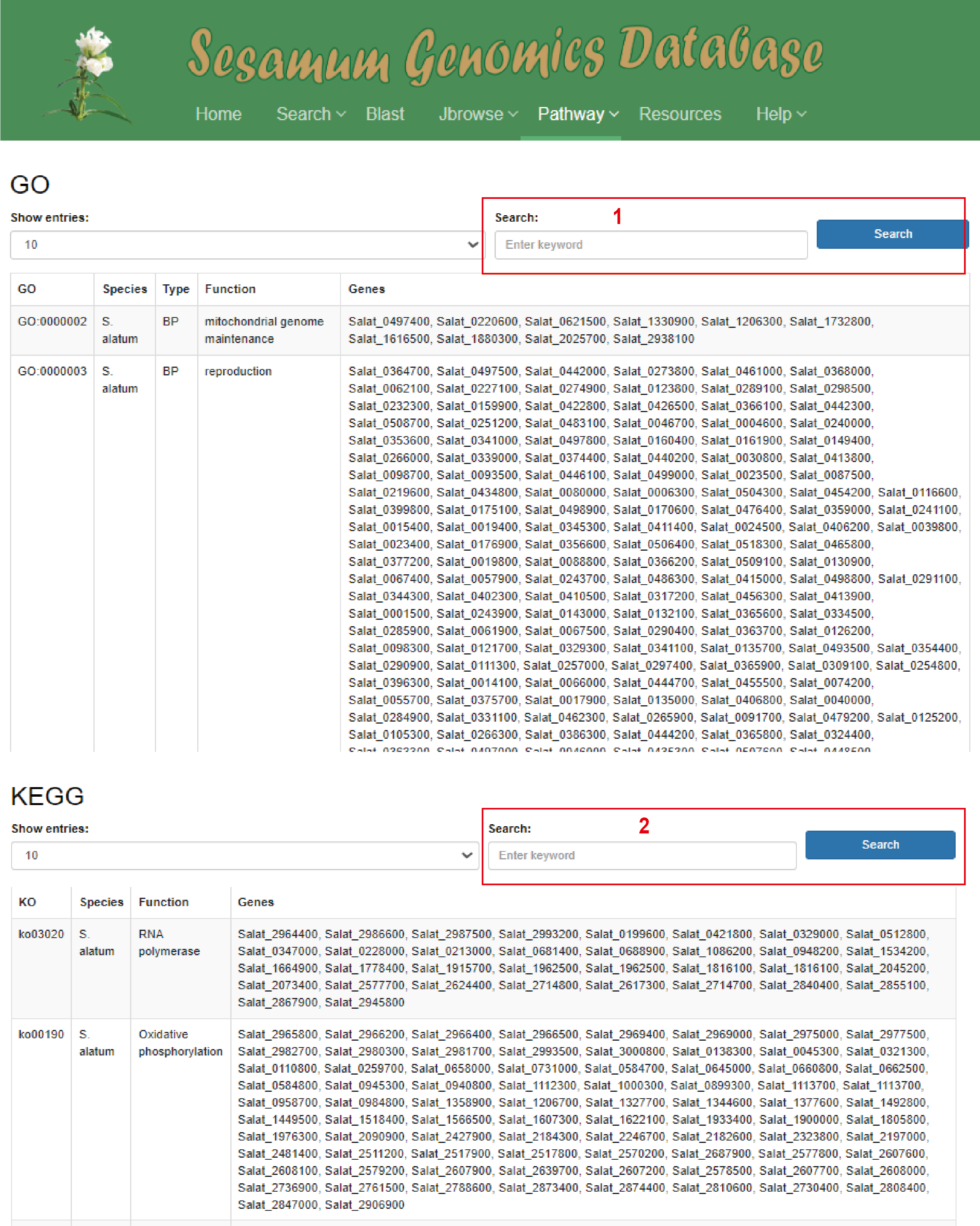
The database offers a multi-search capability for both GO (box 1 in red) and KEGG (box 2 in red) databases . Users can conveniently input a variety of identifiers such as GO numbers, KO numbers, species names, gene functions, and Gene IDs to directly access the information they seek.
The database offers genome, protein, and GFF3 annotation files in a zipped format, simplifying the download process for users interested in specific species.


